E-mail: pr-research*office.hiroshima-u.ac.jp (Please replace * with @)
Stress can impact sleep, focus and more — down to our molecules. Oxidative stress results from an imbalance of oxidants and antioxidants, caused by an increase of free radicals, or reactive oxygen species, that can damage cells and their components. Observed in several diseases, from Parkinson’s disease to hepatitis to cancer, chronic oxidative stress also appears to cause hypoxia, another kind of stress condition involving insufficient oxygen in tissues. But it’s not well understood how stress impacts biological processes at the molecular level.
Researchers from Hiroshima University are a step closer to understanding potential common features of sources of oxidative stress. They analyzed 839 collections of genetic material, called transcriptomes, related to oxidative stress from public databases. From this meta-analysis, they identified 386 pairs of transcriptomes related to oxidative stress. The researchers compared these findings to previously identified samples of hypoxia-related transcriptomes to determine what genes may be involved in both types of stress.
The research team published their results on December 3 in Biomedicines.
Also Read: Scientists use public databases to leap over scourge of publication bias
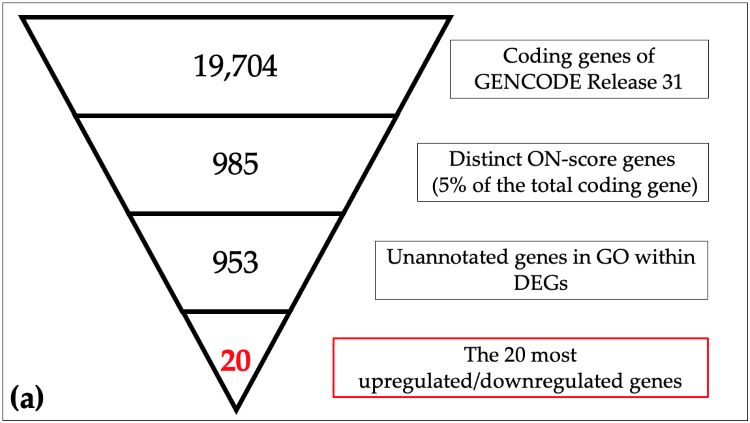
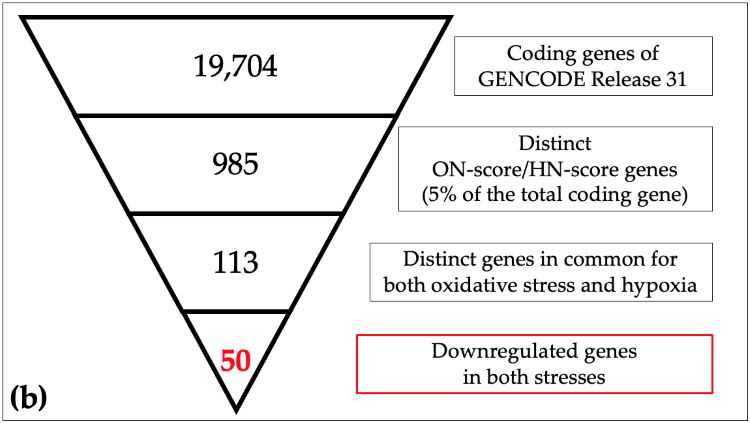
Schematic view of identification of responsive genes in meta-analysis
(a) The 19,704 coding genes indexed for the reference genome were filtered by ON_score and by excluding Gene Ontology (GO) annotated genes to retrieve the 20 most differentially expressed genes (DEGs). (b) The number of genes down-regulated in oxidative stress and hypoxia was then obtained as per the schematic in the figure. (Image courtesy of Takayuki Suzuki, Hiroshima University)
According to paper author Hidemasa Bono, professor in the Graduate School of Integrated Sciences for Life, Hiroshima University, an immense range of external and internal factors can contribute to oxidative stress.
“Distinguishing oxidative and non-oxidative sources is challenging and complicated, so our present study focused on analyzing the common features among various sources of oxidative stress from the perspective of changes in gene expression,” Bono said. “Our meta-analysis identified common factors for oxidative stress and hypoxia in a data-driven way.”
The researchers’ meta-analysis found two genes, called CRIP1 and CRIP3, were particularly downregulated during instances of oxidative stress. According to Bono, this finding suggests a relationship between oxidative stress and zinc homeostasis. The two genes encode proteins that can bind to zinc, and CRIP1 transports and absorbs zinc.
Previous studies have reported several roles for zinc in antioxidant defense systems, including contributions to processes that reduce reactive oxygen species, according to Bono. He also noted that zinc is a known component of superoxide dismutase, a protein that works to reduce and maintain levels of reactive oxygen species in cells.
When the results of the oxidative stress meta-analysis were compared to the results of the hypoxia meta-analysis, the researchers found that several genes were differentially expressed under both stress conditions, meaning the same genes were observed at different levels depending on the condition.
“We inferred that the downregulation of cell cycle-related genes is a mutual biological process in both oxidative stress and hypoxia,” Bono said. “But the common feature among various those oxidative stress sources have not been clearly explained yet. We have not yet defined a clear line between oxidative stress and other types of stress such as hypoxia.”
Although precise molecular mechanisms underlying various sources of stress are still unknown, Bono said that the results of this study may play a role in elucidating the causes and in developing treatments for diseases related to both oxidative stress, hypoxia and zinc homeostasis.
“The results and methods of this research are expected to be an important resource for future research focused on understanding the functions of the responsive genes discovered,” Bono said. “This is because the data-driven nature of meta-analysis makes it possible to obtain new findings that are difficult to achieve with traditional hypothesis-driven research methods.”
According to Bono, the use of collective intelligence, including the datasets and future results of this study in next steps, will make it possible to efficiently promote studies on the search for key pathways, for causes and treatments of diseases.
Contributors include Takayuki Suzuki and Yoko Ono, also with the Graduate School of Integrated Sciences for Life at Hiroshima University.
The Center of Innovation for Bio-Digital Transformation, the Japan Science and Technology Agency (JPMJPF2010) and the Research Organization of Information and Systems’ Joint Support Center for Data Science Research (009RP2021) supported this research.
About the study
Journal: Biomedicines
Title: Comparison of Oxidative and Hypoxic Stress Responsive Genes from Meta-Analysis of Public Transcriptomes
Authors: Takayuki Suzuki, Yoko Ono & Hidemasa Bono
DOI: 10.3390/biomedicines9121830
Norifumi Miyokawa
Office of Research and Academia-Government-Community Collaboration, Hiroshima University

 Home
Home







