E-mail: tetsushi-sakuma*hiroshima-u.ac.jp (Please replace * with @)
Genome editing becomes predictable, scalable, and highly efficient
Japan -- Researchers led by Professor Takashi Yamamoto and Dr. Tetsushi Sakuma report a new genome editing system that brings predictable outcome with high scalability and high efficiency.
The technique, described in Nature Communications, attaches the landing pad for the repair molecules of the DNA breaks at the editing site, which plays a key role to regulate the pattern of the edits.
Genome editing technology has enabled to engineer the genomic DNA of any organisms and cultured cells, in which site-specific DNA double-strand break (DSB) is introduced and repaired via various endogenous repair pathways. The editing outcome is totally dependent on the stochastic pathway choice, resulting in uncertain modifications to be introduced.
To firmly control the repair pathways, followed by the intended edits, scientists at Hiroshima University devise the novel method of recruiting the controller molecule to the target locus in collaboration with Professor Kohichi Tanaka and Dr. Tomomi Aida at Tokyo Medical and Dental University.
Dr. Tetsushi Sakuma, Associate Professor at Hiroshima University and one of the corresponding authors on the study, called the new system LoAD, or Local Accumulation of DSB repair molecules. The LoADing strategy and its application were conceived by Dr. Shota Nakade, the first author on the study, as well as Dr. Sakuma.
“Genome engineers have always worried about technical problems such as unexpected large deletion and low knock-in efficiency, because pinpoint designation of selected DSB repair pathway at the target site was impossible,” said Nakade, “I surmised that the accumulation of DSB repair molecules at the cut site would make these pathways controllable.”
The method of “utilizing” particular repair pathway has been well established. The authors previously developed the PITCh (Precise Integration into Target Chromosome) method, directed against a certain kind of pathways, microhomology-mediated end-joining (MMEJ), to insert the exogenous DNA sequence with good efficiency.
Indeed, the sophisticated design of gene targeting including PITCh has effectively utilized endogenous repair pathway such as MMEJ; however, there apparently was room for improvement in terms of “biasing” the pathway selection, because the way to steer the pathway was not proactively determined.
The major achievement using the LoAD system was the enhancement of gene insertion with the PITCh method, by accumulating MMEJ-triggering molecule, CtIP. CtIP proteins can introduce end resection at the DNA break site, followed by homology-directed repair including MMEJ.
The PITCh-enhancing activity of CtIP reached about 2-fold, suggesting that MMEJ was successfully induced predominantly at the target locus. The effect was confirmed at various gene loci in various cell lines. It clearly showed that the authors successfully put the genome editing machinery on the “right track” to the defined edits.
Based on this, they further showed simultaneous insertion of multiple gene cassettes using the LoADed PITCh strategy. Three distinct cassettes were inserted into the intended target loci independently by single transfection and cloning, without any selection. Thus, the LoAD system improved not only efficiency but also scalability.
“Ultimately, genome editing outcome should be perfectly controlled so that it could produce no by-products. Our LoAD system developed in this study could bias the repair pathway largely, which paved the way for the final goal,” said Dr. Sakuma.
The expanded applications of the LoAD system other than PITCh were also reported in the study. They showed that CtIP accumulation could enhance various types of gene insertion mediated by other repair pathways such as homologous recombination and single-strand template repair. Furthermore, donor-free genome editing such as the introduction of short deletion and multiple mutagenesis could be selectively conducted by LoADing another repair molecule, Trex2. These results suggest that the LoADing strategy can facilitate a wide variety of genome editing operations.
Professor Takashi Yamamoto, a supervisor of the study, has a high hope for the future application and expansion of the LoAD system.
“We became to get a great command of genome editing. LoAD system will enable us to reproduce various types of disease mutations in cells and organisms.”
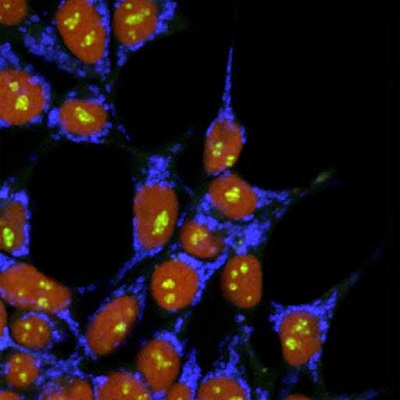
Figure: Multicolored cells established with simultaneous insertion of triple gene cassettes using the LoADed PITCh system (Hiroshima University)
This work was funded in part by grants from the Japan Society for the Promotion of Science (17K15045 to S.N., 16K18478 to T.S., and 17H01409 to T.Y. and T.S.) and the Japan Agency for Medical Research and Development (AMED) (Research Program on Hepatitis to T.Y.).
Full bibliographic information
- Journal: Nature Communications
- Title: Biased genome editing using the local accumulation of DSB repair molecules system
- Authors: Shota Nakade1, Keiji Mochida1, Atsushi Kunii1, Kazuki Nakamae1, Tomomi Aida2,3, Kohichi Tanaka2, Naoaki Sakamoto1, Tetsushi Sakuma1,4, and Takashi Yamamoto1,4
Author Affiliations:
1 Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University
2 Laboratory of Molecular Neuroscience, Medical Research Institute (MRI), Tokyo Medical and Dental University (TMDU)
3 McGovern Institute for Brain Research, Massachusetts Institute of Technology
4 Corresponding authors. - doi: 10.1038/s41467-018-05773-6
- Profile of Associate Professor Takashi Yamamoto
- Profile of Associate Professor (Lecturer) Tetsushi Sakuma
Tetsushi Sakuma
Associate Professor (Lecturer)
Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University


 Home
Home