Reference article: Seirin-Lee S, Osakada F, Takeda J, Tashiro S, Kobayashi R, Yamamoto T, et al. (2019) Role of dynamic nuclear deformation on genomic architecture reorganization. PLoS Comput Biol 15 (8): e1007289. DOI: https://doi.org/10.1371/journal.pcbi.1007289
Original text by Margaux Phares, edited by Emma Buchet
Biological experiments confirm mathematical modeling of retina development in mice

Changes in the shape of DNA structure affect the nuclei of nocturnal animals. These findings in PLOS Computational Biology could help explain how nocturnal animals, such as mice, see at night.
By combining mathematics with science, an interdisciplinary team at Hiroshima University (HU) found how changes in the shape of DNA structure affect the nuclei of nocturnal animals. Their findings could help explain how nocturnal animals, such as mice, see at night.
Nocturnal and diurnal mammals see the same – but only for a brief time. When mice are born, the chromatin in the cells of their eyes has a diurnal structure. Day by day, the layout of this chromatin slowly inverts, allowing the mice to see at night. How this change happens was a mystery.
Sungrim Seirin-Lee, Associate Professor, and Hiroshi Ochiai, Lecturer, in the Graduate School of Integrated Sciences for Life at HU, suspected that the chromatin was making the shape of the nuclei change shape. “When we started this research, our hypothesis was based 100 percent on mathematics,” Seirin-Lee said. “Because of our mathematical modeling, we found that nuclear deformation might be a key point in DNA’s structure change.”
If we could see inside of the nucleus, we would see that chromatin comes in different types and territories. Around the center of the nucleus is euchromatin, or DNA that is largely active. Heterochromatin, on another hand, is a kind of DNA that lies around the envelope or ceiling of the nucleus. Unlike euchromatin, the gene activation of heterochromatin is low.
Between nocturnal and diurnal animals, though, the differences in nuclear architecture get bigger – especially around the retina. The DNA is in the center of the nucleus in nocturnal mammals. Usually, heterochromatin stays put in the nuclear envelope. In the case of nocturnal animals, though, Seirin-Lee and Ochiai found it can be moved by the nucleus changing shape.
To describe the movement of chromatin, Seirin-Lee and her colleagues used a type of mathematical modeling called phase-field modeling. A method commonly used in physics; phase-field modeling can be used to do things like telling apart ice from water. However, according to Seirin-Lee, “it is not common in the biological sciences. In chromatin dynamics, it is the first trial in the world!” Using this function, the group could see the movement of chromatin and nucleus by determining and defining the inside and outside of the nucleus, as well as euchromatin versus heterochromatin.
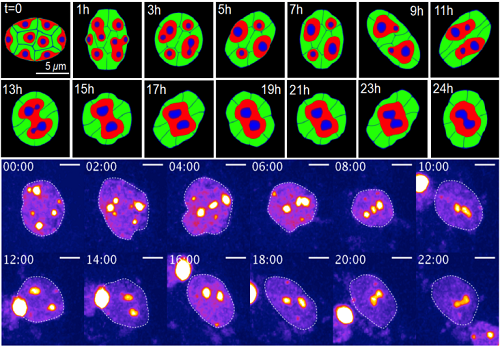
Change of chromatin structure induced by dynamic nuclear deformation (Upper: simulation result by mathematical model, Below: live imaging data of experiment)
When the group observed heterochromatin in the mouse’s eyes, they found that conditional architecture triggered dynamic deformation, which resulted in an inverted nuclear architecture. In the inverted architecture case, two proteins are removed, which allows heterochromatin to move.
Then, with the assistance Ochiai, they put their model to the test on neural stem cells, which mimic retinal cells. After treating the cells with proteins that keep heterochromatin at the nuclear periphery, deformation stopped. Chromatin clustering increased, and nuclear architecture could not finish inverting. This finding was consistent with Lee’s mathematical modeling.
Ultimately, Seirin-Lee and her colleagues want to see if their findings are universal to mammal cells.
“At this stage, we think it is just mouse eyes,” Seirin-Lee said, “but we don’t know! Maybe humans could have such structures by dynamic nuclear deformation.”
Next, Seirin-Lee is looking to tackle the intermediate structure, or a sort of hybrid between conventional and inverted architecture of the nucleus.
This work was supported by JSPS KAKENHI Grant Numbers JP18H05531 and JP19K06612 to H.O.; JP16K17643 to S.S.L; JP15K15631 to F.O., and by JST, PRESTO Grant Number JPMJPR16E2 to S.S.L.; JPMJPR14F6 to F.O.; and JPMJPR15F2 to H.O., Japan.
- Profile of Associate Professor SUNGRIM SEIRIN-LEE
- Profile of Lecturer Hiroshi Ochiai
- Find more Hiroshima University research news on our Facebook page.
Norifumi Miyokawa
Research Planning Office, Hiroshima University
E-mail: pr-research*office.hiroshima-u.ac.jp (Please change * into @)

 Home
Home