E-mail: pr-research*office.hiroshima-u.ac.jp (Please replace * with @)
In recent years, messenger RNA, DNA’s close cousin in life’s complex process of going from a string of genetic blueprints to fully functioning organism, has received intense scrutiny in the scientific and medical community for the role it can play in creating next-generation vaccines, cancer treatments, and stem cell therapies addressing a myriad of previously incurable diseases. The previously obscure topic of mRNA became a nearly universal household utterance following the rush to discover a type of vaccine that could prevent COVID-19 related fatalities. The scientific community’s herculean effort did result in Pfizer’s mRNA COVID-19 vaccine, and products with similar mechanisms of action closely follow from other U.S. and global pharmaceutical companies.
An international research team led by Professor Katsura Asano of Hiroshima University’s Graduate School of Integrated Sciences for Life in Japan, and also of Kansas State University in the U.S., set out to find new ways to artificially induce mRNA to respond in ways that could eventually lead to therapeutic outcomes, expanding on the success of the mRNA-based COVID-19 vaccines and opening up new possibilities across a host of possible genetic therapies.
Asano and his research team paid attention to a biochemical process termed chemical modification that adds a chemical mark to RNA bases, corresponding to a genetic letter of life’s blueprint, and identified such chemical marks that both speed up and slow down action in the beginnings of the chemical zippers involved in generating gene-specified proteins. They published their findings on April 8, 2022 in Science Advances.
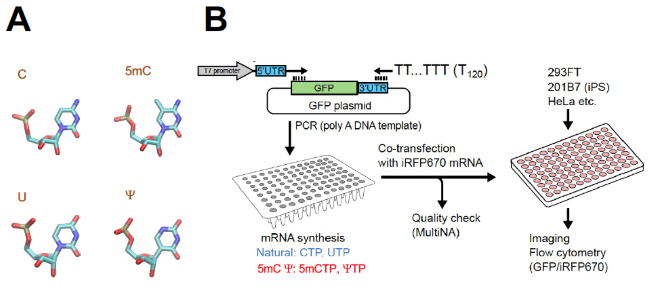
Experimental approaches
A, examples of chemically modified nucleotides used in this work shown to the right. Shown to the left are the natural nucleotide, cytosine (C) or uridine (U) before modification. B, experimental scheme. See legends to Fig. 1A-B of the original paper. (Image courtesy of Katsura Asano, Hiroshima University and Kansas State University)
In animals, including humans, mRNA is called to action in the protein production process with a signal called the AUG Start Codon, a universal code for the genetic “zipper” of RNA. The compound that AUG makes up is an amino acid called methionine, one of the twenty building blocks of protein molecules. Other RNA codons such as GUG (amino acid Valine), UUG (amino acid Leucine), and CUG (also Leucine) are generally considered “non-start” codons, meaning they’re less likely to represent the beginning of a gene translation. Instead, they appear in the middle of protein coding region that is meant to unzip the genetic blueprint and produce a given protein.
Few other codons than AUG are known to be able to activate mRNA in the way AUG does. But in setting out to change that, Asano and his team set out to test common RNA chemical modifications, evaluating their effects on different types of rare start codons initiating the translation process. To do so, they used their previous discovery that GUG, UUG, and CUG codons that are different by one letter from AUG, are converted to a reasonably strong start codon specifying methionine through attaching the optimum RNA sequence for initiating their translation event in animals. Their study design pitted a dozen RNA sequences, derived from these sequences, for expressing green fluorescent proteins through various non-AUG start codons at various efficiencies. To accurately evaluate GFP expression, they used a technique called flow cytometry to measure fluorescence from ~10,000 cells per attached RNA sequence and start codon. In this way, they compared translation efficiencies between natural RNA and chemically modified RNA.
They found common trends in altering translation efficiencies when a certain non-AUG start codon received a certain chemical mark. A remarkable discovery, they reported, was the ability of U-to-Psi (pseudouridine) conversion to dramatically increase initiation potentials of CUG, GUG and UUG start codons (and more satisfyingly no affect on AUG). “Chemical modification of non-AUG start codons can greatly alter initiation frequencies from these codons,” Asano said. “Computer simulation played a key role in understanding the mechanism leading to these effects. mRNA translation from non-AUG start codons is an old but new concept. These start codons were used in prokaryotes [bacteria] but our research takes the concept a big step further by highlighting the possibilities of doing so in eukaryotes, including humans.”
Asano hopes the medical industry will take note of this new body of data and continue to conduct further research into how to use chemical modified RNA for generating synthetic expression switches—in such a way to stimulate translation activity in a highly targeted way in humans and animals. “I am hoping that the companies making mRNA vaccines will use our findings,” he said. “For example, they could use UUG start codon and chemically modify mRNA by 1m Psi, as Pfizer did with their COVID-19 vaccine. They will allow strong expression of the antigen from the start codon and yet avoid protein expression from cDNA made and integrated into genome by chance.”
Asano explained further that so far, no significant risks related to long-term use of various mRNA vaccines have been identified. “But there is a small chance that vaccines against retroviruses make vaccine cDNA when the patient encounters these viruses during immunization. If this integrates into the patient’s genome, the antigen may be expressed in a way that attenuates vaccine production for boosting,” he said. “But beyond that, the concept is so easy and adds no extra cost. So we hope these techniques are adopted.”
###
Asano is also affiliated with the Hiroshima Research Center for Healthy Aging, Hiroshima University.
About the study
Journal: Science Advances
Title: Translational recoding by chemical modification of non-AUG start codon ribonucleotide bases
Authors: Yoshihiko Fujita, Takeru Kameda, Chingakham Ranjit Singh, Whitney Pepper, Ariana Cecil, Madelyn Hilgers, Mackenzie Thornton, Izumi Asano, Carter Moravek, Yuichi Togashi, Hirohide Saito & Katsura Asano
DOI: 10.1126/sciadv.abm8501
Norifumi Miyokawa
Office of Research and Academia-Government-Community Collaboration, Hiroshima University

 Home
Home