Research
In our laboratory of molecular nutrition, education and research are conveyed in molecular and genetic levels in priority of physiological functions of nutrients and food factors. In recent years, a number of people who have obesity, diabetes, and cancers has been increasing with westernization of meal, and it has become a big social problem. Thus, the goal of our laboratory is to elucidate nutrients and food factors preventing these illnesses as well as their action mechanisms.
To date, the nucleotide sequence of the human genome has been completely determined. Owning to this, the mechanisms of human diseases and the functions of nutrients and food factors are understood at a gene level. In our laboratory, we have worked on elucidation of the mechanisms underlying lifestyle-related diseases, such as obesity, diabetes, and hypertension, through cell culture studies, gene expression analysis, and animal experiments by using transgenic and gene knockout mice, etc. These studies could provide the molecular targets for disease prevention. Discovery of such molecular targets will be tremendous useful for novel evaluation of food functions and development of functional foods in the future.
Keywords
Molecular nutrition, Nutrients, Food factors, Life-style diseases,Molecular targets
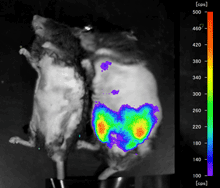
A non-invasive way to assess the anti-inflammatory properties of functional foods against obese adipose tissue
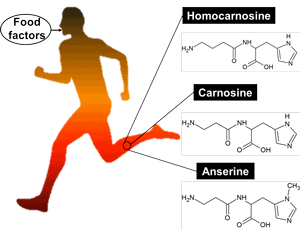
Searching for food factors increasing imidazole peptides in skeletal muscle
Recent Publications
Nirmagustina DE, Yang Y, Kumrungsee T, Yanaka N, Kato N. (2018) Gender Difference and Dietary Supplemental Vitamin B6: Impact on Colon Luminal Environment. J. Nutr. Sci. Vitaminol., 64, 116-28.
Yang B, Kumoto T, Arima T, Nakamura M, Sanada Y, Kumrungsee T, Sotomaru Y, Shimada M, and Yanaka N. (2018) Transgenic mice specifically expressing amphiregulin in white adipose tissue showed less adipose tissue mass. Genes Cells. 23, 136–145.
Mitsumoto K, Watanabe R, Nakao K, Yonenaka H, Hashimoto T, Kato N, Kumrungsee T, and Yanaka N. (2017) Time-course microarrays reveal early activation of the immune transcriptome in a choline-deficient mouse model of liver injury. Life Sci. 184, 103-111.
Sanada Y, Yamamoto T, Satake R, Yamashita A, Kanai S, Kato N, van de Loo FA, Nishimura F, Scherer PE, Yanaka N. (2016) Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages. Sci. Rep. 6, 38697.
Hashimoto T, Yang B, Okazaki Y, Yoshizawa I, Kajihara K, Kato N, Wada M, Yanaka N.(2016) Time Course Analysis of Skeletal Muscle Pathology of GDE5 Transgenic Mouse. PLoS One. 11, e0163299.
Ohshima N, Kudo T, Yamashita Y, Mariggiò S, Araki M, Honda A, Nagano T, Isaji C, Kato N, Corda D, Izumi T, Yanaka N. (2015) New members of the mammalian glycerophosphodiester phosphodiesterase family: GDE4 and GDE7 produce lysophosphatidic acid by lysophospholipase D activity. J. Biol. Chem. 290, 4260-4271.
Kumrungsee T, Wang ZQ, Matsumura S, Saiki T, Tanaka M, Matsui T. (2014) Identification of peptides from soybean protein, glycinin, possessing suppression of intracellular Ca2+ concentration in vascular smooth muscle cells. Food Chem. 152, 218-224.
Okazaki Y, Ohshima N, Yoshizawa I, Kamei Y, Mariggiò S, Okamoto K, Maeda M, Nogusa Y, Fujioka Y, Izumi T, Ogawa Y, Shiro Y, Wada M, Kato N, Corda D, Yanaka N. (2010) A novel glycerophosphodiesterphosphodiesterase, GDE5, controls skeletal muscle development via a non-enzymatic mechanism. J. Biol. Chem. 285, 27652-27663.


 Home
Home
