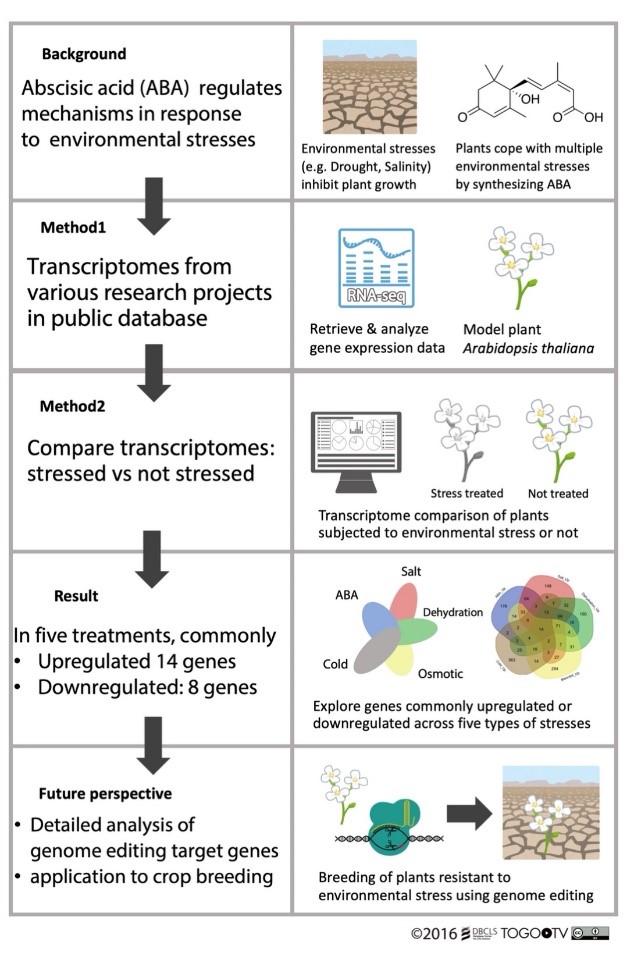
To better understand the molecular pathways that allow ABA to increase, the research team analyzed public RNA sequencing data on thale cress, or Arabidopsis thaliana. RNA sequencing is a technique that enables scientists to identify and quantify specific sequences of genetic instructions programmed in an organism’s RNA. This data can reveal how different variables may increase or decrease the expression of certain genes.
Bono and his team specifically focused on five ABA-related stress conditions: ABA, when the hormone is applied directly to the plant; salt, which changes how the plant can use water; dehydration, or how much water the plant has; osmotic, when plant cells swell or shrink inappropriately; and cold.
“The data-driven studies have the advantage of analyzing large and independent datasets, which can lead to the identification of novel targets, distinct from the extensively studied established factors and accelerate the development of stress-tolerant crops,” Bono said.
The researchers performed a meta-analysis of 216 paired datasets, combining those research results and reanalyzing them to identify where data might overlap or reveal previously unknown connections.
The meta-analysis revealed that 14 genes were commonly up-regulated and eight genes were commonly down-regulated across all five ABA-related stress responses investigated. Bono noted that some genes regulated by salt, dehydration and osmotic treatments were not regulated by ABA or cold stress, suggesting that they may be involved in the plant response through a different signaling pathway.

 Home
Home