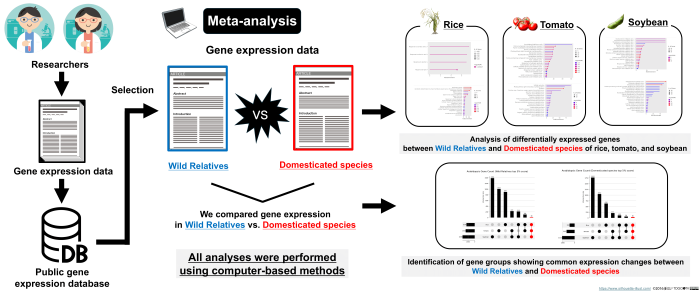
This diagram illustrates the methodology of a meta-analysis comparing gene expression data between wild relatives and domesticated species. Using public gene expression databases and computer-based methods, researchers analyzed data from rice, tomato, and soybean to identify differentially expressed genes and common expression changes associated with domestication. (Makoto Yumiya / Hiroshima University, CC BY 4.0)
With climate change and more frequent extreme weather events, researchers predict that global yields of important crops like maize, rice, and soybeans could decline by 12 to 20% by the end of the century. To prepare, plant scientists are hoping to find ways to improve yields and grow hardier varieties of these crops. New insights into the genetic makeup of wild varieties of common crops show how domestication has changed crop traits over time and propose a new cultivation method to improve genetic diversity. The research was shared in a paper published in Life on July 11.
“While domesticated species have originally been bred by cultivating wild species, the resulting reduction in genetic diversity can damage all individuals by exposing them to diseases and environmental stresses. To solve this problem, we set out to identify differences in crop traits between wild relatives and domesticated species and to contribute to the selection of new breeding candidate genes. The introduction of useful traits, especially those found in wild relatives, may provide hints for the development of new useful varieties,” said Hidemasa Bono, a professor at Hiroshima University’s Graduate School of Integrated Sciences for Life in Hiroshima, Japan.
The researchers used RNA sequencing data from public databases, including the National Center for Biotechnology Information Gene Expression Omnibus and published studies online. They focused on crops with wild relatives that had widely available RNA sequencing data: tomatoes, rice, and soybeans. The gene expression data of the wild varieties was then compared to the domesticated varieties. To evaluate the data, researchers classified all genes into three groups: upregulated, unchanged, and downregulated.
By understanding the gene expression comparison between the wild varieties and the domesticated varieties, researchers could understand differences in how the plants respond to stressors. “Wild relatives have high environmental stress tolerance with the potential to respond to climate change and severe changes in the natural environment, which has been an issue in recent years,” said Bono.
The researchers found 18 genes that were upregulated in the wild relatives and 36 genes that were upregulated in the domesticated species. Wild species were found to have genes related to environmental stress responses while domesticated species had more genes related to the hormone regulation and chemical compound export and detoxification. For example, a gene called HKT1 affects salt stress response and salt tolerance was found to be upregulated in wild varieties. This could be an opportunity to develop crops that can grow in soil with more saline. Researchers also found genes that were upregulated in wild varieties that helped with drought stress (RD22), water stress (HB-12), leaf development and photosynthesis promotion (HB-7), and osmotic stress response and wound signaling (MYB102).
In domesticated plants, researchers also found beneficial genes that were upregulated compared to wild varieties. Several genes help detox the plants and remove chemicals found in soil. One gene (ALF5) improves the plant’s resistance to tetramethylammonium, and another (DTX1) manages cadmium and toxic compounds. These genes and others can help plants grow in soils that have been contaminated by chemicals. Researchers suspect this may have become beneficial for plants because of increased pesticide and chemical fertilizer use.
“The three wild species used in this analysis—rice, tomato, and soybean—had in common high expression levels of genes that contribute to stress responses, such as drought, osmotic pressure, and wound stress. The high expression levels of genes that contribute to stress tolerance that these three less closely related species have in common suggest that wild species of other species are likely to have useful traits as well,” said Bono.
Looking ahead, researchers hope to learn even more about these essential differences between wild relatives and domesticated species to improve breeding. “In addition, we would like to collect and reanalyze data used in crop breeding research to construct a database that will contribute to the promotion of digital breeding of crops,” said Bono.
The other contributor to this research was Makoto Yumiya of the Graduate School of Integrated Sciences for Life at Hiroshima University.
The Center for Bio-Digital Transformation (BioDX), COI-NEXT, and the Japan Science and Technology Agency (JST) supported this research.
This paper received funding from Hiroshima University to cover open access fees.

 Home
Home