By Caitlin E. Devor
Originally Published October 20, 2016
Mathematical analysis has led researchers in Japan to a formula that can describe the movement of DNA inside living human cells. Using these calculations, researchers may be able to reveal the 3D architecture of the human genome. In the future, these results may allow scientists to understand in detail how DNA is organized and accessed by essential cellular machinery.
Previous techniques of studying the genome's architecture have relied on methods that require killing the cells. This research project, involving collaborators at multiple institutes in Japan, used alternative molecular and cell biology techniques to keep the cells alive and collect data about the natural movement of DNA.
DNA is often envisaged as a stable and static code, but the genome as a whole is actually an active molecule that moves around and changes shape. Currently, scientists can sequence the entire basic code of DNA, but knowing the larger-scale 3D architecture of the genome would reveal more information about how cells use the code.
While the cell is growing, DNA is stored as an unraveled spool of string; certain portions (euchromatin) are more loosely wound, and therefore accessible to the cellular machinery that turns DNA into protein, than other areas that are kept tightly wound (heterochromatin). When the cell prepares to split in half during cell division, it packages all of the chromatin into tightly-wound, X-shaped chromosomes.
"Our calculations consider the fractal dimensions of the DNA, which shows how densely the DNA is packed inside the cell. The way the DNA is packed may indicate how the cell uses certain genes," said Soya Shinkai, PhD, Assistant Professor at Hiroshima University and first author of the research paper.
Along the "string" of chromatin are regularly-spaced, barrel-shaped "beads" of DNA-protein complexes called nucleosomes. Researchers tracked nucleosomes' movement around the cell to understand where and how chromatin is stored.
Researchers labeled the nucleosomes with fluorescent tags and took microscopy images during the growth phase of human cells. They then used theories of polymer physics to quantify the movement of the nucleosomes.
"Every second, a 10 nanometer-sized nucleosome can move 100 nanometers. The constant, subtle random forces within the cell make the chromatin move around so much," said Shinkai.
Before a cell can use a gene, the DNA must be completely unwound. Areas of chromatin containing frequently used genes are less tightly wrapped than areas of chromatin with infrequently used genes. A model to visualize how chromatin is packed within the cell could allow researchers to understand which genes are accessed most or least often and how the genome is physically organized.
"Our calculations are relevant to local chromatin structures, but this method could also be extended to whole chromosomes. These mathematical formulas are a theory for how to interpret the visual data from microscope images of DNA moving within the cell," said Yuichi Togashi, PhD, Associate Professor at Hiroshima University and last author of the research paper.
Future research projects will include finding new microscopy and DNA labeling techniques to visually track the movement of individual nucleosomes over longer periods time.
The four researchers who published the recent paper are experts in theoretical and computational biophysics, structural biology, cell biology, and molecular biology. The results come from a research project made possible by an interdisciplinary team of scientists associated with the Research Center for the Mathematics on Chromatin Live Dynamics at Hiroshima University. Other collaborators for the project include the National Institute of Genetics (Japan), Keio University, and Sokendai Graduate University for Advanced Studies.
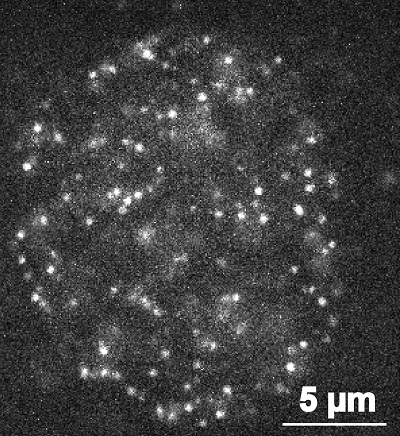 |
| Image of a human nucleus, the center of a cell where DNA is stored. |
IMAGE CAPTION: White spots are nucleosomes, microscopic structures of short length DNA-protein complexes. Researchers collected visual data about the movement of nucleosomes in living human cells and used the information to calculate the 3D architecture of the genome. Images like this one were part of an interdisciplinary project by researchers at Hiroshima University, the National Institute of Genetics (Japan), Keio University, and Sokendai Graduate University for Advanced Studies.
IMAGE CREDIT: Image by Tadasu Nozaki and Kazuhiro Maeshima, originally published in PLOS Computational Biology.
IMAGE LICENSE: Image is available under the Creative Commons Attribution license (CC-BY) and may only be re-used with attribution to T. Nozaki, K. Maeshima, and PLOS Computational Biology.
Full Citation:
Authors: Shinkai S, Nozaki T, Maeshima K, Togashi Y
Title: Dynamic nucleosome movement provides structural information of topological chromatin domains in living human cells
Journal: PLOS Computational Biology (20 October 2016)
DOI:10.1371/journal.pcbi.1005136
Media Contact:
Norifumi Miyokawa
Research Planning Office, Hiroshima University
pr-research*office.hiroshima-u.ac.jp (Please replace * with @)

 Home
Home














